Most of us would be aware of the prognostic value of the immunoglobulin heavy-chain variable region gene (IGHV) mutation status of our CLL (See healthunlocked.com/cllsuppo... for the history.) In general, IGHV mutation status has the greatest influence on our journey with CLL. Roughly 50% of us diagnosed with CLL are fortunate enough to be IGHV mutated and are thus likely to have a long period in watch and wait, so have a greater hope of falling into the group of around 30% of us who never need treatment. Those of us with unmutated IGHV CLL have most benefited from the recent revolution in CLL treatments away from chemoimmunotherapy 'chemo', to targeted treatments. It's the immunoglobulin structure in B Cell Receptors (BCRs) that enable memory B cells to quickly detect and respond to threats to our health and change to the mature plasma cell B cell stage, where they become immunoglobulin manufacturing factories. CLL cells appear a lot like memory B cells; they rely on signalling through their BCRs to stay alive. Without that signalling, they eventually die, which is why BTK inhibitors, the 'brutinibs' and PI3K inhibitors, the 'lisibs', are so effective at controlling CLL.
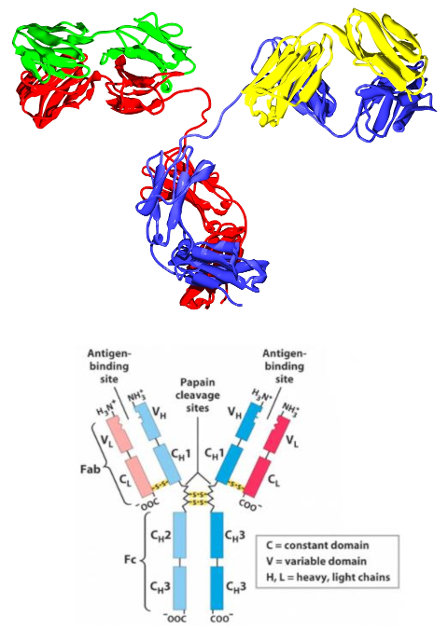
The accompanying image shows the protein structure of IgG and the stylised representation with which we are most familiar. Our immune system relies on a process termed somatic hypermutation to change the DNA we inherit - our germ line DNA, so that B cells can match the billions of different proteins found in invaders and recognise them for neutralisation and destruction. Somatic hypermutation is the purposeful change to our B cell DNA by which a major part of our adaptive immune system functions, rather than an unwanted cancer causing mutation. Basically, by deliberately mutating our B cell DNA to randomly change the amino acids used to make the heavy and light chain proteins in the immunoglobulin molecule, some B cells in our nodes will key with the invader's proteins and go on to produce plasma cells and memory B cells.
That long introduction brings me to the reason for this post, this article in Nature, brought to my attention by CLLerinOz nature.com/articles/s41375-...
From the abstract, with my emphasis;
The mutational status of immunoglobulin (IG) light chain genes in chronic lymphocytic leukemia (CLL) and its clinical impact have not been extensively studied. To assess their prognostic significance, the IG light chain gene repertoire in CLL patients has been evaluated using a training-validation approach. In the training cohort (N = 573 CLL), 92.5% showed productive IG light chain genes rearrangements, with IGKV4-1 (20.5%) and IGLV3-21 (19.0%) being the most common. A 99.0% somatic hypermutation cut-off was identified as the best predictor for time to first treatment (TTFT) in 414 Binet A CLL patients of the training cohort. Patients with unmutated (UM) light chain genes displayed a 10-year treatment free probability of 32.4% versus 73.2% for those with mutated (M) genes (p < 0.0001). Importantly, UM light chain genes maintained an independent association with a shorter TTFT when adjusted for the IPS-E prognostic model variables, that also includes IGHV mutational status. The validation cohort of 343 Rai 0 patients confirmed these findings, with UM light chain genes predicting a 7-year treatment free probability of 42.0% versus 73.7% for M genes (p < 0.0001). These results indicate that the mutational status of the light chain genes is an independent predictor of shorter TTFT in early-stage CLL patients.
This was a relatively large study for CLL research and the study group was deemed representative of newly diagnosed patients;
The patient characteristics of the training cohort (n = 573) at the time of diagnosis were consistent with a real-world cohort of unselected newly diagnosed CLL (Table 1), since the median age at diagnosis was 70 years, 58.8% were male and 41.2% were females, the median lymphocyte count was 9800/μl, 35.8% had unmutated (UM)-IGHV, 43.5% had del, (13q) 8.0% harbored del (17p) and 9.8% presented TP53 mutations.
To what extent these study results will later be included in regular prognostic testing and beyond, is yet to be seen, but it does further explain why CLL is such a heterogeneous illness.
As I mentioned earlier, the revolution in CLL treatment to targeted therapy came out of research into what could be done to switch off (inhibit) B Cell Receptor signalling. That revolution has resulted in the approval of ibrutinib, acalabrutinib, zanubrutinib and pirtobrutinib, with many more BTKi drugs also in clinical trials, per this post healthunlocked.com/cllsuppo...
PI3K inhibitors, such as idelalisib, duvelisib, umbralisib, zandelisib, et al, also covered in the above post, have unfortunately been less successful.
Our antibody levels and their impact on our ability to fight of infections, are also of relevance, because of how CLL suppresses plasma cell antibody production. That's why hypogammaglobulinemia (low IgA, IgG, IgM) is fairly common when we have CLL and unfortunately tends to worsen over time.
For those interested in further reading about antibodies/immunoglobulins;
Antibody Structure, classes and functions (the lower structural image of IgG is derived from this reference).
antibodysystem.com/archive/...
The IgG molecule visualisation is from the Wikimedia collection commons.wikimedia.org/wiki/...
Antibodies are immune system-related proteins called immunoglobulins. Each antibody consists of four polypeptide – two heavy chains and two light chains joined to form a "Y" shaped molecule.
The amino acid sequence in the tips of the "Y" varies greatly among different antibodies. This variable region, composed of 110-130 amino acids, give the antibody its specificity for binding antigen. The variable region includes the ends of the light and heavy chains.
The structure of a typical antibody molecule
ncbi.nlm.nih.gov/books/NBK2...
Antibodies are the secreted form of the B-cell receptor. An antibody is identical to the B-cell receptor of the cell that secretes it except for a small portion of the C-terminus of the heavy-chain constant region. In the case of the B-cell receptor the C-terminus is a hydrophobic membrane-anchoring sequence, and in the case of antibody it is a hydrophilic sequence that allows secretion. Since they are soluble, and secreted in large quantities, antibodies are easily obtainable and easily studied. For this reason, most of what we know about the B-cell receptor comes from the study of antibodies.
Structure and characteristics of antibody isotypes
Human antibodies are classified into five isotypes (IgM, IgD, IgG, IgA, and IgE) according to their H chains, which provide each isotype with distinct characteristics and roles.
ruo.mbl.co.jp/bio/e/support...
Immunoglobulin vs Antibody: Unveiling the Intricacies
assaygenie.com/blog/immunog...
In the realm of immunology, the terms "immunoglobulin" and "antibody" often find themselves used interchangeably, sparking confusion among those keen on understanding the immune system's fine details. While closely related, these terms encapsulate nuances vital for a comprehensive grasp of how the body defends itself against pathogens.
:
While every antibody is an immunoglobulin, not every immunoglobulin serves as an antibody. This distinction hinges on the function and specificity of the immunoglobulin in the immune response.
Neil