Further to healthunlocked.com/cllsuppo...
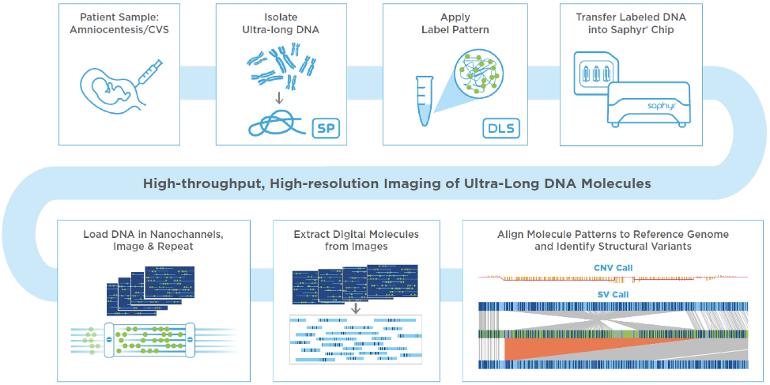
An update from watching the OGM webinar
My impression is that AML cells' DNA has greater structural variation (mutation) than seen in CLL. Some of the combinations of deletion/ duplication/ insertion/ translocation described in AML were mind-boggling, and occasionally there occur "cryptic" translocations, invisible to FISH, nevertheless clinically significant. OGM can achieve 20 to 100 times the resolution of other analytical methods.
That said, it seems likely that when OGM is routinely applied to CLL, genomic variations will be discovered that were not apparent before, enabling certain "complex karyotypes" to be better understood and new prognostic markers to be identified. We have a glimpse of this already in the recent paper ncbi.nlm.nih.gov/pmc/articl... and the earlier ashpublications.org/blood/a...
The OGM workflow is straightforward compared with karyotyping plus FISH/ NGS etc. The claim that OGM "could potentially replace current cytogenomic methodologies" (see abstract above) was not made in the webinar. It was more a case of: we know OGM excels at certain things, it doesn't excel at everything, how can we use it alongside other methods? An international working group has been set up to to examine this question, whiich is also considered in a recent Special Report in Blood journal ashpublications.org/blood/a...
Wider applications of Optical Genome Mapping
OGM has been used to identify rare genetic mutations (the non-pejorative term is structural variations) that may pre-dispose people to progressing to severe Covid19 sciencedirect.com/science/a...
OGM for the chromosomal characterization of cell lines used in preclinical and clinical research aacrjournals.org/cancerres/...
A clinical trial to detect genomic structural variants in human DNA by OGM compared to prior clinical genetic tests such as chromosomal microarray analysis (CMA), karyotyping, Southern blot analysis, polymerase chain reaction (PCR), fluorescence in situ hybridization (FISH), and/or next generation sequencing (NGS), etc. in 1,000 individuals with an identified genomic abberration clinicaltrials.gov/ct2/show...