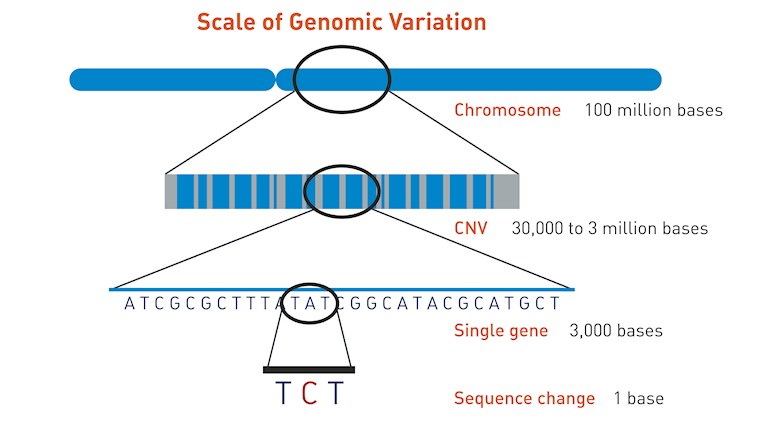
Chromosomal microarray (CMA) is clever, very sensitive technology which looks for extra (duplicated) or missing (deleted) chromosomal segments, sometimes called copy number variants (CNVs) - see illustration. These include: microdeletions and microduplications of chromosome segments, which are too small to see under a microscope but may contain multiple genes, most abnormalities of chromosome number (trisomy, monosomy, etc.) and most unbalanced rearrangements of chromosome structure (translocations, etc.)
CMA testing is often used to help the families of children with previously undiagnosed conditions but now it's also being used to establish clonality and high risk features in patients with Richter Transformation where it's helpful in tracking duplications or deletions of chromosomal segments, including aberrations involving TP53 and CDKN2A (a key regulator of cell cycle progression), which are known to be markers of poor prognosis.
This poster from ASH 2019 looks at 24 patients with a diagnosis of Richter transformation to DLBCL with a prior history of CLL/SLL. Of these patients, 7 had CMA performed on both the CLL/SLL and RT samples.
CMA was able to provide proof of common clonality in 6 of 7 patients with Richter transformation to DLBCL, which is a poor prognostic feature of RT. It also identified a loss of TP53 and/or CDKN2A in 5 of 7 patients, also a poor prognostic feature of RT.
"This information can be used to counsel patients on prognosis and could effect clinical recommendations such as treatment with allogeneic stem cell transplantation. Institutions with the ability to run CMA should utilize this modality in patients Richter transformation to DLBCL."
Full details of the poster here: ash.confex.com/ash/2019/web...
Jackie