This paper is not very helpful to us in terms of advancing treatment. But it might be of interest and help in understanding what goes on.
It is all too easy to read, then repeat terms like "receptor" and yet have next to no visualisation of what they are!
Or you can scroll through, view the pictures, and go "Wow!" 
Being a review paper it does paint a more complete picture than the highly specific research papers. But the language does make some of it difficult to properly appreciate.
Structural Insights Into Thyroid Hormone Receptors
Endocrinology, Volume 166, Issue 1, January 2025, bqae154, doi.org/10.1210/endocr/bqae154
Abstract
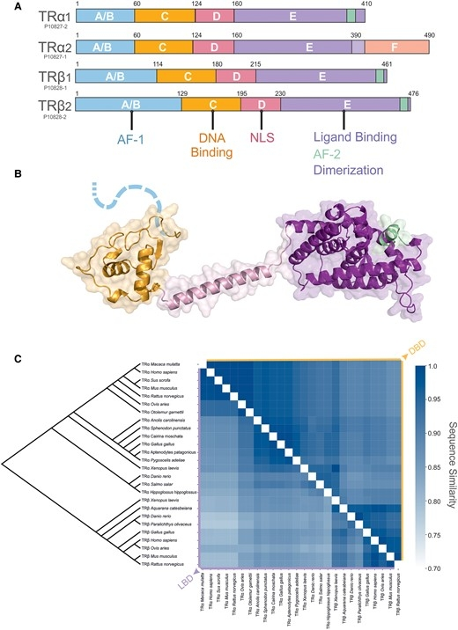
Thyroid hormone receptors (TRs) are essential components of the endocrine system, mediating the cellular effects of thyroid hormones. The 2 TR genes, THRA and THRB, encode 4 isoforms, with TRα1 and TRβ1 being the most prevalent. TRs are ligand-dependent transcription factors and members of the nuclear receptor superfamily, indispensable for human growth, development, and metabolism. Dysfunctional TR signaling can lead to conditions such as resistance to thyroid hormone (RTH) syndrome, thyroid cancer, and metabolic disorders. Structurally, TRs comprise several domains: a variable N-terminal domain, a conserved DNA-binding domain, and a ligand-binding domain that mediates interaction with hormones and transcriptional coregulators. TRs predominantly function as heterodimers with the retinoid X receptor (RXR), binding to thyroid hormone response elements in target genes to regulate their transcription. This review examines the structural studies on TRs, primarily performed through x-ray crystallography, that have provided detailed insights into TR functions, including DNA recognition, ligand binding, and coregulator interactions. We also discuss how these findings have deepened our understanding of TR mechanisms and contributed to the interpretation of pathogenic mutations.
Keywords: thyroid hormone receptor, atomic structure, DNA recognition, ligand binding, coregulator interaction, pathological mutation
Open access: