A fine example of evolutionary principles applied to metastatic PCa with the development of a method (Mscore) for risk assessment based on key gene mutations. (U of Texas and Chinese U collaboration.)
* * *
Abstract
Background Metastasis is the primary cause of prostate cancer-related deaths. However, the underlying molecular mechanisms and evolutionary patterns remain largely uncharacterized.
Methods We evaluate the heterogeneity and genomic evolution of prostate cancer with multi-organ metastases. The samples include 32 primary samples, 23 lymph node metastases, 22 bone metastases, 16 liver metastases, and four pelvic mess metastases. They are analyzed to identify the mutated genes enriched in metastatic samples, selected by metastases, and leading to different long-distance migrations. These metastasis-related alterations constitute a Mscore for evaluating the metastatic risk of primary prostate tumors.
Results Our analysis discovers 21 metastasis-related mutated genes in total. Of them, 14 genes are finally selected for metastatic risk prognosis, including the mutations of AR and KMT2C with high prediction ability. A Mscore established with these 14 characteristics by the xgboost model displays its ability to classify primary tumors and metastases. This score can further divide primary prostate tumors from the TCGA cohort into two groups. The two subsets present significantly differential survival risks. This score can also identify metastasis-featured primary tumors for breast cancer, bladder cancer, liver cancer, and uterine corpus endometrial carcinoma.
Conclusion Our research proposes 14 molecular features potentially driving prostate cancer metastasis. The Mscore established on them can estimate the metastatic risk of primary tumors.
* * *
Of special note is the following from the Results Section of the paper:
* * *
3 Results
3.1 The heterogeneity between primary and metastatic prostate tumors
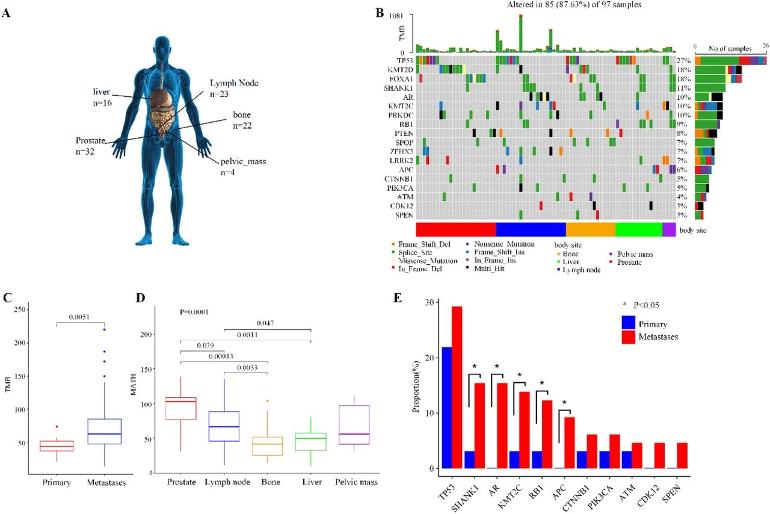
Our pipeline identified 248 somatic single nucleotide variants (SNVs) and 163 somatic copy number variants (CNVs) across the tumors of 72 prostate cancer patients. Of the genes with somatic SNVs, the most frequently mutated ones were TP53 (27%), KMT2D (18%), FOXA1 (19%), SHANK1 (11%), AR (10%), and KMT2C (10%) (Figure 1B). Of the somatic CNVs in Figure S2, the most frequent CNVs included losses at 17q21.31, 16q22.1, and 6q15 regions and gains at 8q24.3 and 12q13.2 regions. These results were consistent with the analyses conducted in previous studies (Figure S3) [25-28]. The analyses confirmed the reliability of our pipeline.
The analysis of these variants revealed that the metastatic samples had high tumor mutational burden (TMB, Figure 1C) and low tumor heterogeneity (MATH, Figure 1D). It showed newly occurred and similar mutations when tumors metastasized to liver, bone, and other organs. The comparisons of the variants between primary tumors and metastases also provided 11 metastasis-enriched features (Figure 1E). They included mutated TP53, SHANK1, AR, KMT2C, RB1, APC, CTNNB1, PIK3CA, ATM, CDK12, and SPEN. Specifically, AR mutations were rare in primary prostate tumors but common in metastatic prostate cancer. KMT2C mutations were significantly high in metastatic samples. Combined with previous studies [26, 29], these results suggested the 11 features as potential metastatic drivers. All the above results revealed the genomic heterogeneity between primary and metastatic prostate tumors.
Moreover, genetic differences existed not only between primary tumors and metastases but also among diverse distant sites of metastases from prostate tumors. Specifically, the MATH value of lymph node metastases was the highest compared to other metastatic sites (Figure 1D). This observation may indicate that a portion of the metastases involved lymph nodes as nodes of dissemination. And the lymph node metastases had more variants (Figure S4), suggesting a higher degree of genomic instability and molecular alterations contributing to the metastatic process through lymph nodes. Additionally, the gene of AR was frequently mutated in lymph node metastases (22%, Figure S4B) and bone metastases (18%, Figure S4C). It may describe the potential roles of AR mutations in the progression of bone and lymph node metastases. These results highlighted the diverse mutational profiles and genetic heterogeneity across different metastatic sites. (emphasis added)
* * *
Full paper link:
Heterogeneity and genomic evolution of metastatic prostate cancer, bioRxiv Preprint, Posted September 04, 2023.
biorxiv.org/content/10.1101...
Foundations for precision medicine are all around us. Unfortunately, not yet in the clinic. Most of us will be history by the time it gets there.
Saluti - CaptnMojoe