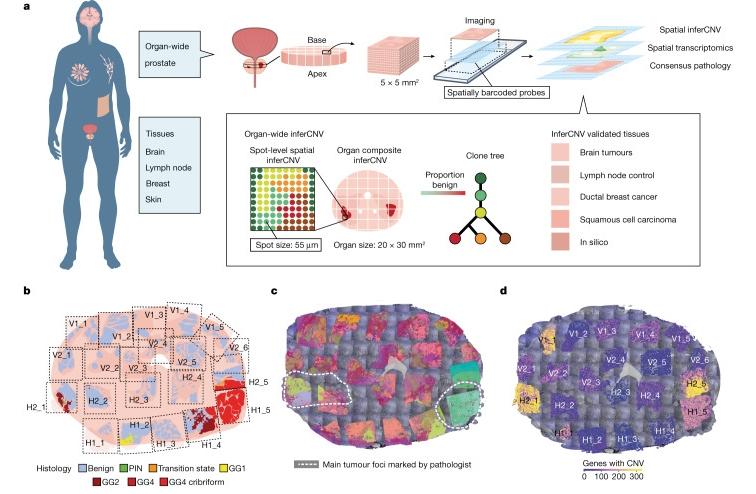
Here is presented the ultimate biopsy of the prostate with an amazing array of discoveries about, and confirmation of, the heterogeneous nature of prostate cancer = the identification of 50K tissue domains within a single patient and 120K across 10 patients.
Below is the discussion section from this complex research paper. BTW, "CNV"= copy number variations
* * *
Discussion
We show that spatial transcriptomic data across multiple cancer types can robustly be used to infer CNV , as validated by FISH and WGS. Specifically, we performed an in-depth spatial analysis of the prostate organ that generated an unprecedented atlas of up to 50,000 tissue domains in a single patient and 120,000 tissue domains across ten patients. For these domains, we inferred genome-wide information in each spot, which facilitated data-driven clone generation in a tissue-wide fashion at high resolution. Notably, the spatial information allowed us to identify small clonal units not evident from morphology, which would therefore be overlooked by histologically guided laser microdissection or even random sampling of single cells. We go on to show that, in some tumour types, particularly in prostate, glioma and breast cancers, CNV analysis identifies distinct clonal patterns within tumours, in line with a recent spatial genome methodology that has also shown granularity in the study of multiclonality of tumours28.
Focusing on prostate cancer, the patterns, as defined by the conservation of CNVs across morphological entities, indicate hitherto unappreciated molecular relationships between histologically benign and cancerous regions. It is known that CNVs occur early in tumorigenesis21. We propose that CNVs can precede tumorigenesis and are a feature of glandular morphogenesis, with propagation of particular variants traversing disease pathology. It seems that clonal status alone and the copy number alterations described here retained in heritable clonal lineages at cell division are insufficient to deliver immediate phenotypic transformation. We believe that our work generates interesting hypotheses regarding epigenetic determinism29 and the environmental effect with, for example, the stromal niche or cross-talk between neighbouring clones. Furthermore, questions remain about the timing of events and how long is needed for morphological transformation to occur. Expression analysis of altered benign clones identified changes consistent with enhanced phenotypic versatility, suggesting that these cells may represent an intermediate state between benign and malignant cells—metabolically active as they try to survive the mutational burden they have acquired, before phenotypic transformation. In summary, this study shows that CNVs in regions of the genome that encode certain cancer drivers (for example, MYC and PTEN) are truly early events, occurring in tissue regions currently unknown to and therefore ignored by pathologists (Extended Data Fig. Fig.4d).4d). This is important given that the risk stratification delivered by pathologists dictates to a large degree treatment decisions and subsequent clinical outcome.
Our study therefore provides an unbiased avenue to interrogate genomic integrity, adding to the armamentarium of cancer molecular pathology. Our findings provide a basis for improved early detection of clinically important cancers, targeted focal and systemic therapy, and improved patient outcomes for ubiquitous malignancies such as prostate cancer. Overall, our study raises important biological questions about cancer evolution, somatic mosaicism and tissue development.
(emphasis added)
* * *
Full paper is here:
Spatially resolved clonal copy number alterations in benign and malignant tissue - Nature. 2022; 608(7922): 360–367. Published online 2022 Aug 10
ncbi.nlm.nih.gov/pmc/articl...
Complexity writ very large. Stay Well - Ciao - K9 terror