For those wanting to understand the science "foundation" for liquid biopsies, this research paper provides a detail account of the techniques, methods, and results for one of the leading companies providing them. It was partially funded by FoundationOne and prepared by their scientists, so the research is specific to their product.
From the paper:
Discussion
Tumor tissue-based testing has historically been the standard of care for genomic analysis, but in many situations adequate tissue sampling is not possible, thus the role of liquid biopsy has rapidly evolved as another option for initial genomic testing. Liquid biopsy also offers opportunities to explore the underlying evolving tumor in a minimally invasive way to help inform cancer management. Additionally, due to intratumor heterogeneity, a tumor biopsy may represent a small sample of the overall tumor cell population, a limitation that can potentially be overcome with liquid biopsy [10, 16–18]. While, it has been recognized that patients could benefit from a cfDNA-based CGP assay (liquid biopsy) with proven performance [13], concern has been expressed around the robustness and lack of performance data available for cfDNA-based genomic tests [27].
The FoundationOne Liquid CDx panel is based on the gene content of the FoundationOne®CDx assay, the first FDA-approved tissue-based broad companion diagnostic that is analytically and clinically validated for all solid tumors. The ability of the FoundationOne Liquid CDx assay to also robustly detect genomic alterations demonstrated in tissue to predict response to targeted therapies has been demonstrated in this study.
The analytical performance of FoundationOne Liquid CDx was evaluated across >7,500 tests and >30,000 unique variants over >300 genes and >30 cancer types, allowing for a comprehensive assessment of performance. Across >900 variants the high sensitivity of the assay was demonstrated with a median 95% LoD of 0.40% VAF in the enhanced sensitivity region and 0.82% VAF in the standard sensitivity region for short variants; 0.37% VAF and 0.90% VAF in the enhanced and standard sensitivity regions, respectively, for rearrangements; 21.7% TF for copy number amplifications; and 30.4% TF for copy number losses. Through the analysis of >130,000 variants, the variant detection rate in healthy donors was determined to be 0% for rearrangements and CNAs and 0.013% (~1 in 8000) for short variants (substitutions and indels). The high reproducibility of variant calling was demonstrated with 99.59% reproducibility across more than 1 million data points. Across >900 positive variants and >150,000 negative variants, an overall PPA of 96.3% and NPA of >99.9% was observed when comparing to an orthogonal cfDNA-based NGS method. Together, these data demonstrate robust genomic analysis results from the test. The analytic performance demonstrated for the FoundationOne Liquid CDx assay is comparable to other NGS-based broad molecular profiling liquid biopsy assays [28–30].
The body of evidence of regarding correlation between liquid biopsy results and patient response to therapy is growing [31–36]. For example, the clinical utility for the selection of patients for therapy with alectinib using an early version of the Foundation Medicine liquid biopsy assay has also been demonstrated in a prospective clinical study described in the Blood First Assay Screening Trial (BFAST) [31].
Critically, clinical validity, particularly through bridging studies, provides confidence in genomic profiling results for cancer patients. In this study, we also describe the clinical validation of the FoundationOne Liquid CDx assay, including large-scale comparisons with orthogonal clinical plasma- and tissue-genotyping methods, for both EGFR in NSCLC and PIK3CA in breast cancer. In 2016, the Roche cobas EGFR Mutation Test v.2 became the first US FDA approved cfDNA companion diagnostic for patients with NSCLC. The performance comparison between FoundationOne Liquid CDx assay and this orthogonal assay for detection of EGFR exon 19 deletions and exon 21 L858R alterations demonstrated non-inferiority concordance.
The clinical bridging study for the second US FDA approved liquid biopsy-based companion diagnostic test demonstrated a 55% PPA (95% CI 49.0, 60.1) and 97% NPA (95% CI 94.0, 99.0) between tissue and liquid results [37]. A similar performance comparison approach between tissue and liquid was utilized in the FoundationOne Liquid CDx validation studies. The clinical validity of the FoundationOne Liquid CDx assay to identify breast cancer patients harboring PIK3CA alterations eligible for treatment with alpelisib was assessed through retrospective testing of plasma samples. In this bridging study, all available plasma samples from patients collected at baseline prior to randomization into the SOLAR-1 clinical trial [24] were tested with FoundationOne Liquid CDx, with results compared to tissue genotyping performing using the SOLAR-1 CTA. The PPA and NPA between FoundationOne Liquid CDx and the tissue-based CTA assay were 71.7% (95% CI 65.4%, 77.5%) and 100% (97.2%, 100%), respectively. Most notably, the clinical efficacy of alpelisib in combination with fulvestrant for the FoundationOne Liquid CDx-positive population was demonstrated with an estimated 54% risk reduction in disease progression or death in the alpelisib plus fulvestrant arm compared to the placebo plus fulvestrant arm (HR = 0.46, 95% CI: 0.30, 0.70).
The number of targeted therapies and actionable alterations continues to grow in many tumor types, including breast cancer [13]. While PIK3CA alterations occur in 36% or more of breast cancer patients [38] there are other relevant biomarkers to consider in these patients, including ERBB2, BRCA1, BRCA2, NTRK, and MSI (or mismatch repair deficiency) [13]. Furthermore, targetable alterations ae increasingly being identified across numerous other solid tumor types, including non-small cell lung cancer [39], prostate cancer [40], colorectal and other gastrointestinal malignancies [41], ovarian cancer [42], melanoma [43], and others. The studies herein describe the ability to interrogate and identify alterations in these relevant genes in a single assay, providing pertinent genomic information from a single, minimally invasive molecular test.
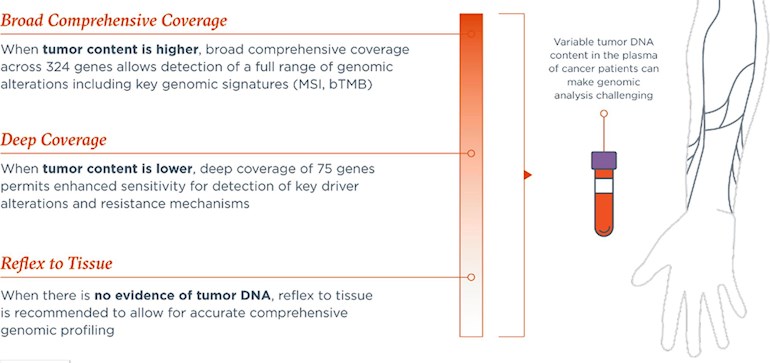
Throughout an individual cancer patient’s journey, liquid and/or tissue testing may be most appropriate to identify genomic alterations indicative of response or resistance to therapy. While the utilization of a robustly validated liquid biopsy assay has definite advantages, including minimally invasive blood sampling and providing a more comprehensive representation of the patient’s entire tumor burden, the variable extent of tumor shedding into the plasma and resultant ctDNA levels can make genomic analysis challenging. Thus, the availability of well-aligned CGP-based tumor- and liquid-based testing assays can maximize benefits to patients. For patients with a high ctDNA content, broad comprehensive coverage across the targeted 324 gene FoundationOne Liquid CDx panel allows detection of a full range of genomic alterations, including key genomic signatures, including MSI and bTMB. For those patients where ctDNA content is lower, deep coverage of 75 genes permits enhanced sensitivity for the detection of key driver alterations and resistance mechanisms. In cases where no ctDNA is detected, reflex to tissue testing is recommended, if possible, to allow for accurate comprehensive genomic profiling to enable data-driven treatment decisions.
The results of the extensive studies presented here demonstrate that FoundationOne Liquid CDx accurately and reproducibly detects the major types of genomic alterations as well as complex biomarkers, such as MSI, bTMB, and tumor fraction. The data described here support the validity and utility of using a well-validated cfDNA-based CGP assay in the therapeutic management of cancer patients. The comprehensive nature of the described performance evaluation studies demonstrates the reliability of test results which can provide confidence to physicians in the use of this assay to provide genomic profiling results for their patients. (emphasis added)
Full paper is here:
journals.plos.org/plosone/a...
And here is the excellent source article referenced (40) in above research:
Towards precision oncology in advanced prostate cancer
ncbi.nlm.nih.gov/pmc/articl...
Don Pescado's Science just keeps on coming.
So, while we all patiently wait for it to get here, Keep Staying Safe & Being Well. Captain K9