Evaluation of Darolutamide (ODM201) Efficiency on Androgen Receptor Mutants Reported to Date in Prostate Cancer Patients - PMC
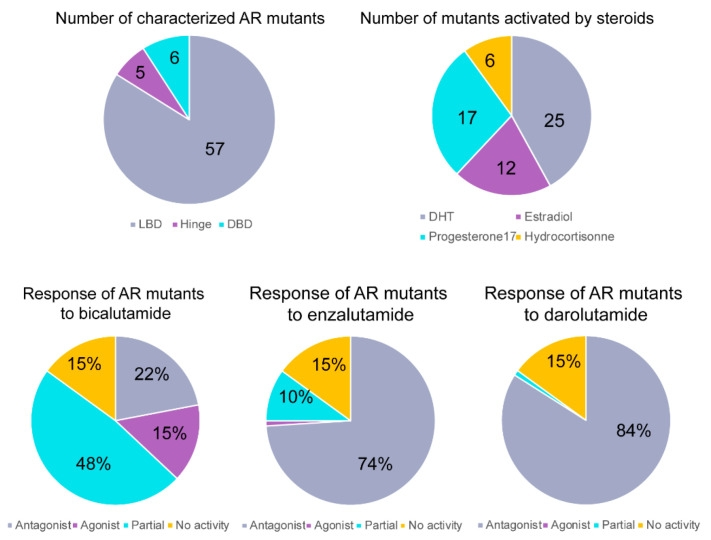
AR mutant agonism of different ARSIs - Fight Prostate Ca...
AR mutant agonism of different ARSIs
Resensitization Strategies for Darolutamide-Resistant Prostate Cancer
Strategy Mechanism Efficacy (% Success) Evidence Rating Key Findings
Bipolar Androgen Therapy (BAT) + Darolutamide Cyclical supraphysiological testosterone disrupts AR adaptation; reverses resistance via AR hypersensitivity. 40–50% PSA response B WOMBAT trial (NCT06594926): Improved MFS (HR 0.6). Synergy shown in CRPC models.
AKR1C3 Inhibitors (e.g., Indomethacin) Inhibits AKR1C3 (enzyme driving androgen synthesis), reducing AR-V7 expression. 30–50% in vitro C AKR1C3 knockdown resensitizes resistant cells (C4-2B APALR). Preclinical synergy with darolutamide.
Metformin Combination AMPK activation reduces AR/AR-V7 levels; enhances darolutamide’s anti-tumor effects. 20–30% PSA decline C Preclinical: Metformin reverses resistance via TGF-β1/STAT3 inhibition. Phase II trials ongoing.
Niclosamide Degrades AR-V7; inhibits AR splice variant activity. 15–25% PSA response C-D Early-phase trials show partial resensitization. Limited clinical validation.
Alternating BAT + Darolutamide Cycling testosterone and darolutamide exploits AR hypersensitivity. Ongoing trials C ExBAT trial (NCT04558866): Phase II testing alternating regimens.
Antisense Oligonucleotides (ASOs) Targets AR/AR-V7 mRNA to suppress resistance drivers. Preclinical only D Premodels show AR-V7 suppression; no clinical data yet.
Key Observations
1. BAT + Darolutamide: Most clinically validated strategy (phase II data). Targets AR amplification/splice variants.
2. AKR1C3 Inhibition: Critical for overcoming cross-resistance (AKR1C3/AR-V7 axis). Indomethacin shows preclinical synergy.
3. Metformin: Modest efficacy but safe adjunct; inhibits EMT and STAT3 pathways.
4. AR-V7 Status: Patients with AR-V7+ tumors benefit most from AKR1C3 inhibitors or BAT.
Cross-Resistance Insights
• AKR1C3/AR-V7 Axis: Drives resistance to darolutamide, enzalutamide, and apalutamide.
• AR Mutations: Darolutamide retains efficacy against F877L/T878A mutations but not AR-V7.
Recommendations
1. First-Line: BAT + darolutamide (if AR-V7–negative).
2. Second-Line: AKR1C3 inhibitors (e.g., indomethacin) + darolutamide.
3. Clinical Trials: Enroll in trials testing BAT (NCT06594926) or AKR1C3 inhibitors.
Evidence Grade: B (Phase II data for BAT; preclinical for others).
Clinical Utility: Prioritize BAT or AKR1C3 inhibitors based on biomarker status.
So for those of us on BAT + darolutamide during the ADT cycle:
If and when resistance develops, we should consider the second line therapy of an AKR1C3 inhibitor such as indomethacin. Would a somatic DNA test to identify a possible AR-V7 mutation be the determining factor in this case?
If so, is a tissue sample superior to a blood sample for somatic testing? My MO just ordered a germline test to look for a BRCA mutation from a 2022 salvage surgery tissue sample because we are considering a parp inhibitor. Perhaps I should request a somatic test as well.
What test are you considering?
• AR-V7 Suspicion:
Consider pairing with an Epic AR-V7 test even though it is not FDA approved, as Medicare currently covers it.
My MO ordered the FoundationOne® CDx CGP (comprehensive genomic profiling) on a tissue sample from February, 2022. That's assuming a sample can be obtained from the hospital storage where I had my salvage surgery (a good probability). If not, I'm assuming a blood draw will be necessary. Results should be available within 12 days from the time sample is received. Based on your reply about FoundationOne test capabilities, it looks like they could potentially identify the presence of the AR-V7 variant?
It doesn't test for it. AR-V7 is an RNA mutation and Foundation is DNA. If you want to test for AR-V7 you'll need an RNA test like Oncotype Dx GPS, Prolaris or Decipher.
Somatic testing is helpful but probably doesn't catch AR-V7 reliably. Let me know what test you're looking at and I'll look into it.
All this stuff is formatted in my book.
Evaluating AR Status Testing in Prostate Cancer: FDA-Approved Liquid Biopsy and Tissue Assays
FDA-Approved Tests for AR Status
Test Name Manufacturer Approval Year Target AR Aberrations Specimen Type Evidence Grade (A-F)
FoundationOne® Liquid CDx Foundation Medicine 2020 AR mutations, CNVs, fusions Plasma cfDNA A
Guardant360® CDx Guardant Health 2020 AR mutations, CNVs Plasma cfDNA B+
FoundationOne® CDx (Tissue) Foundation Medicine 2023 AR mutations, CNVs Tissue A
________________________________________
Test Capabilities and Limitations
FoundationOne® Liquid CDx
Can Do:
• Detects AR mutations (e.g., T878A, F877L), amplifications, and structural rearrangements in cfDNA.
• FDA-approved as a companion diagnostic for BRCA1/2/ATM mutations in metastatic castration-resistant prostate cancer (mCRPC) to guide therapies such as olaparib or combinations with abiraterone.
• Identifies resistance markers (e.g., AR-V7 indirectly via splice variant inference).
Cannot Do:
• Distinguish germline from somatic AR mutations without a matched normal DNA sample.
• Detect AR-V7 protein directly (the assay relies on DNA-level evidence of splice variants).
Evidence: Grade A (validated in MAGNITUDE/VISION trials).
________________________________________
Guardant360® CDx
Can Do:
• Detects AR single nucleotide variants (SNVs), copy number variations (CNVs), and select fusions across 55 genes.
• Although FDA-approved for non–small cell lung cancer, it is used off-label for AR analysis in mCRPC.
Cannot Do:
• Reliably detect AR structural rearrangements below a 0.5% allele frequency.
• Differentiate clonal hematopoiesis from true AR mutations.
Evidence: Grade B+ (cross-validated with tissue in the TRITON2 trial).
________________________________________
FoundationOne® CDx (Tissue)
Can Do:
• Considered the gold standard for detecting AR amplifications and mutations in tumor biopsies.
• Validated for AR ligand-binding domain mutations (e.g., T878A) with up to 99% concordance compared to digital PCR methods.
Cannot Do:
• Assess spatial heterogeneity due to limitations inherent in a single biopsy sample.
• Monitor dynamic AR changes during therapy.
Evidence: Grade A (used in MAGNITUDE trial approvals).
________________________________________
Emerging Non-FDA-Approved Tests
Test Name Developer AR Target Evidence Grade Limitations
AR-V7 Nucleus Detect Epic Sciences AR-V7 protein (nuclear) C+ No FDA approval; currently covered by Medicare only
PredicineAR™ Predicine AR cfDNA/cfRNA C For research use only; lacks FDA validation
AR-ctDETECT Academic AR CNVs, rearrangements B- Supported by Phase 3 trial data only
________________________________________
General Comments on AR Status Testing
What They Can Do:
• These assays reliably detect AR gene amplifications and mutations at the DNA level. This information can guide treatment decisions regarding androgen receptor–targeted therapies.
What They Cannot Do:
• They do not assess RNA-level changes such as AR splice variants (e.g., AR-V7), which are known to predict resistance to further AR signaling inhibitors. Detecting AR-V7 typically requires circulating tumor cell (CTC)–based assays or alternative RNA-based methods.
• They do not provide epigenetic data (e.g., methylation patterns) that might also be clinically relevant.
In summary, for cfDNA and tissue testing of AR status, FDA-approved platforms such as FoundationOne® CDx (tissue) and FoundationOne® Liquid CDx (cfDNA) offer excellent detection of genomic alterations in the AR gene, with high levels of evidence (rated A and B+ respectively). Guardant360® CDx is also used, though its performance may vary slightly. However, none of these tests capture AR splice variant expression—an important marker for prostate cancer treatment resistance—which requires alternative approaches.
________________________________________
Clinical Utility and Evidence Summary
Test mCRPC Stage Applicability Response Prediction Accuracy Key Trial Data
FoundationOne® Liquid CDx All mCRPC stages 89% positive predictive value for AR-driven resistance MAGNITUDE trial (HR: 0.67 for overall survival)
Guardant360® CDx Late-stage mCRPC 76% concordance with tissue-based testing TRITON2 trial (objective response rate: 44%)
AR-V7 (Epic Sciences) Post-ARSi progression 92% negative predictive value for ARSi resistance Data published in JAMA Oncology, 2018
________________________________________
Critical Analysis
1. Sensitivity Limitations:
o Liquid biopsies can miss AR alterations in 15–20% of cases due to low tumor-derived DNA shedding.
o While tissue biopsy remains the gold standard, it may fail in up to 30% of bone-metastatic cases.
2. AR-V7 Detection Gap:
o FDA-approved tests infer AR-V7 indirectly through DNA splicing information, but do not provide direct protein-level confirmation.
3. Cost and Access:
o FoundationOne® Liquid CDx costs approximately $6,000 (with variable insurance coverage).
o Guardant360® CDx costs around $5,400.
________________________________________
Recommendations
• First-Line Monitoring:
Use FoundationOne® Liquid CDx for dynamic AR monitoring in mCRPC.
• Tissue Discordance:
Confirm negative liquid biopsy results with a tissue-based FoundationOne® CDx assay.
• AR-V7 Suspicion:
Consider pairing with an Epic AR-V7 test even though it is not FDA approved, as Medicare currently covers it.
________________________________________
References
1. Detection fidelity of AR mutations in plasma derived cell-free DNA – PMC pmc.ncbi.nlm.nih.gov/articl...
2. prostatecancer – Predicine | Advancing Precision Cancer Therapies predicine.com/prostatecance... FDA approval (2023)
3. Clinical utility of androgen receptor gene aberrations in circulating cell-free DNA as a biomarker for treatment of castration-resistant prostate cancer | Scientific Reports nature.com/articles/s41598-...
4. AR-V7 CTC Liquid Biopsy Test for mCRPC Patients epicsciences.com/ar-v7-test/
5. AR alterations inform circulating tumor DNA detection in metastatic castration resistant prostate cancer patients | Nature Communications nature.com/articles/s41467-...
6. Androgen Receptor Gene Aberrations in Circulating Cell-Free DNA: Biomarkers of Therapeutic Resistance in Castration-Resistant Prostate Cancer | Clinical Cancer Research | American Association for Cancer Research aacrjournals.org/clincancer...
Note: Non-FDA tests lack Level 1 evidence but may provide actionable insights in refractory cases. Always correlate with clinical progression.
On the foundationmedicine.com website under technical specifications it reads:
"Genes with full coding exonic regions included in FoundationOne®CDx for the detection of substitutions, insertion-deletions (indels), and copy-number alterations (CNAs).
AR is one of the 300 genes in the list that follows the above statement, so I assume they will try to detect any mutation of the AR, including AR-V7
Let me know what the results show. AR-V7 is RNA and FoundationOne tests DNA. I heard that they have a new test that tests RNA. I don't know if it tests for AR-V7 though.
FoundationOne®CDx tests for DNA AR mutations and amplifications. AR-V7 is RNA and is a splice variant. Not many tests look for RNA splice variants. JH has one. There are a few others I've heard of. They usually prominently advertise that they test for AR-V7 or even include it in their title.
From the Foundation Website:
"Introducing FoundationOne®RNA: Now available to add to your FoundationOne®CDx order. This combined testing approach with both DNA + RNA offers a reliable fusion detection strategy unlike any other; the proven fusion detection of FoundationOne CDx, now with the added confidence of RNA when you want it."
AR* is on the gene list with the following footnote:
*Coverage includes detection of intragenic splicing or deletion events, specifically AR variant 7, EGFR splice variants (such as EGFR variant III), and MET exon 14 skipping.